Part of the Flavonoids Compound Library were tested for antiviral activity on SARS-CoV-2 in a cell based assay
Recently, Part of the Flavonoids Compound Library of ChemFaces was purchased by the University of Warsaw, and the literature was published recently. The inhibitor structures were analyzed for structure-activity relationships in order to define common structural patterns. Finally, the most potent inhibitors were tested for antiviral activity on SARS-CoV-2 in a cell based assay.
Among them, ChemFaces has 8 products has antiviral activity on SARS-CoV-2 in a cell based assay.
CFN98599
CFN96488
CFN90835
CFN90960
CFN99571 (
CFN99740
CFN98504
CFN99569
Copyright: University of Warsaw
Source: https://chemrxiv.org/engage/chemrxiv/article-details/60c75800337d6cb114e2912d
Abstract
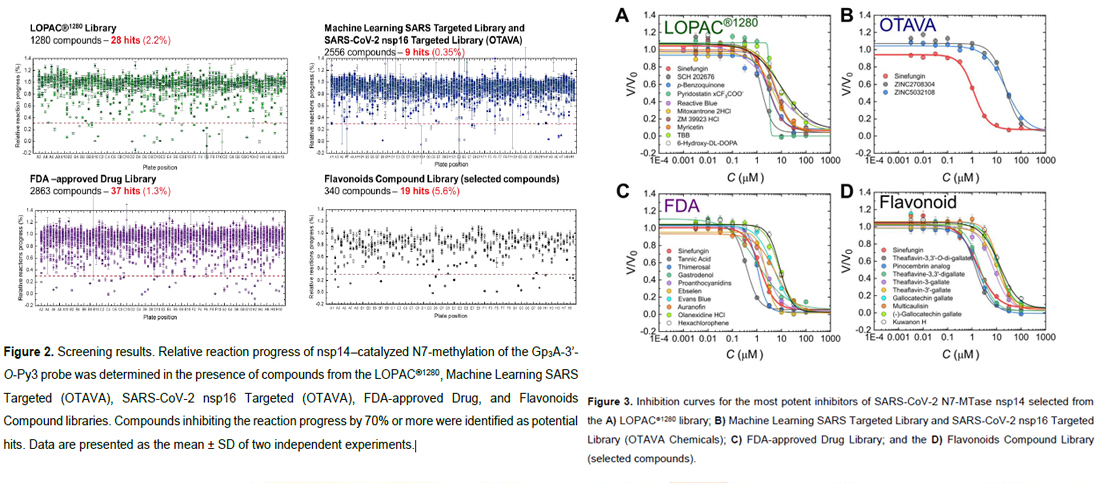
SARS-CoV-2, the cause of the currently ongoing COVID-19 pandemic, encodes its own mRNA capping machinery. Insights into this capping system may provide new ideas for therapeutic interventions and drug discovery. In this work, we employ a previously developed Py-FLINT screening approach to study the inhibitory effects of compounds against the cap guanine N7-methyltransferase enzyme, which is involved in SARS-CoV-2 mRNA capping. We screened five commercially available libraries (7039 compounds in total) to identify 83 inhibitors with IC50 < 50 μM, which were further validated using RP HPLC and dot blot assays. Novel fluorescence anisotropy binding assays were developed to examine the targeted binding site. The inhibitor structures were analyzed for structure-activity relationships in order to define common structural patterns. Finally, the most potent inhibitors were tested for antiviral activity on SARS-CoV-2 in a cell based assay
Experimental Section
General Information. Synthesized compounds were purified by using Flash chromatography on Biotage® Selekt System with different columns and UV-detection at 260 nm or semipreparative HPLC. Yields were calculated based on either sample weight or (preferably) optical milliunits (opt.mu) of the product. Optical unit measurements were performed in 0.1 M phosphate buffer pH 7 at 260 nm. Analytical RP HPLC was performed on Agilent Tech. Series 1200 using Phenomenex Gemini® NX-C18 3 μm HPLC column (150 x 4.6 mm, flow rate 1.0 mL/min) with linear gradient of methanol in 0.05 M ammonium acetate buffer (pH 5.9). UV-detection was performed at 254 nm. Semipreparative RP HPLC was performed on the same apparatus equipped with Gemini® 5 μm NX-C18 110? column (250 x 10 mm, flow rate 5 mL/min) with linear gradient of acetonitrile in 0.05 M ammonium acetate buffer (pH 5.9). UV-detection was performed at 254 nm. The
14
structure and homogeneity of final product was confirmed by RP HPLC, high resolution mass spectrometry HRMS (ESI-), 1H NMR, 1H-1H COSY, 13C NMR, 1H-13C HSQC NMR, 31P NMR, and 1H-31P HMBC NMR spectroscopy. Mass spectra were recorded on Thermo Scientific LTQ Orbitrap Velos (high resolution) and Sciex QTRAP 3200 (low resolution) spectrometers. Routine NMR spectra were recorded at 25 °C and nucleotide concentration of 1–5 mM on a Bruker AVANCE III spectrometer at 500.24 MHz (1H, 1H-1H COSY), 126.80 MHz (13C), and 202.5 MHz (31P). The 1H chemical shifts were determined relative to external standard sodium 3-trimethylsilyl-[2,2,3,3-2H4]-propionate (TSP), and the 31P chemical shifts relative to external standard 20% H3PO4. The nsp14 protein was expressed and purified at the Laboratory of Bioorganic Chemistry, Centre of New Technologies, University of Warsaw, Poland. The RNMT-RAM protein was expressed and purified at the Centre of Gene Regulation and Expression, School of Life Sciences, University of Dundee, Dundee, UK. Solutions of S-adenosyl-?-methionine (SAM) were purchased from New England BioLabs. LOPAC®1280 library of pharmacologically active compounds was purchased from Sigma Aldrich. Machine Learning SARS Targeted Library and SARS-CoV-2 nsp16 Targeted Library were purchased from OTAVA Chemicals. FDA-approved Drug Library was shared by prof Marcin Drag from Wroclaw University of Science and Technology, Poland. Part of the Flavonoids Compound Library was purchased from ChemFaces.